Bacterial Adaptation in Neonatal Meningitis
Despite being uncommon, a significant number of infants with neonatal meningitis do not survive beyond their first month of life. Bacterial meningitis leads to inflammation of the lining of the brain and spinal cord. Neonatal meningitis-causing Escherichia coli (NMEC) is the culprit in one in five cases of neonatal meningitis and is even more common in premature babies.
The gram-negative bacteria NMEC must first cross the blood-brain barrier and steer clear of the brain’s immune cells to cause inflammation and meningitis. This is no easy feat, as immune cells are constantly surveying brain tissue to detect any factors that would compromise the brain’s health.
A study by Sun et al. describes how NMEC bacteria can alter their flagella’s structural components to evade capture by immune cells. Flagella are whip-like structures responsible for bacterial movement. These findings aid the search for therapeutic strategies for bacterial meningitis.
Activation of immune cells by NMEC bacteria
The study1 revealed microglia (or microglial cells) are the first type of immune cell to be activated upon infection with NMEC. Based on data from in vitro experiments, popular belief has been that upon activation, microglia engulf bacteria like NMEC. Surprisingly, analysis using single-cell RNA sequencing techniques (scRNA-seq) and immunofluorescence in this study indicated that only a small proportion of microglia engulf NMEC bacteria. Instead, microglia appears to produce cytokines such as tumor necrosis factor-alpha (TNFα).
Astrocytes are activated to produce complement proteins
Microglia-produced TNFα enhances the ability of a second type of immune cell — astrocytes—to produce complement protein (complement component 3), which can act to lyse and kill bacteria.
Prior to the study, research showed that TNFα production relies on FliC, an important flagellar filament structural protein. This protein interacts with the Toll-like receptor protein (TLR5) on microglia and NFκB signaling. Researchers tested FliC with TLR5 inhibitor and StressMarq’s caffeic acid phenethyl ester (CAPE) (catalog# SIH-263) which is a NFκB inhibitor. Results indicated that inhibition of these factors decreased TNFα levels from bacteria-activated microglia.
NMEC bacteria decrease flagellin synthesis to hide from astrocytes
NMEC bacteria are very sensitive to their environment. NMEC displayed the ability to sense low levels of the amino acid threonine in cerebrospinal fluid (CSF). In these conditions, NMEC reduce
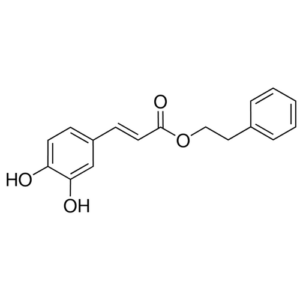
Chemical structure of CAPE (NFκB Inhibitor) (Catalog # SIH-263)
the expression of the flagellin proteins in the flagella of the bacteria that induce astrocytes to produce C3. NMEC bacteria achieve this by using a small RNA (sRNA) to regulate the mRNA levels of flagellin. Consequently, this leads to the downregulation of the production of TNFα by microglia and astrocyte C3, resulting in NMEC survival.
Overall, the study describes for the first time the cascade of the immune mechanism underlying infection by the pathogenic NMEC bacteria. It also shows how the bacteria cleverly adapt to alter their flagellin structures and avoid detection by this immune response cascade. The research delineates how NMEC evade the host brain, potentially providing insights for therapeutic strategies.
StressMarq’s NFκB inhibitor, CAPE, aids research into NMEC infection and other diseases
The described study highlights the use of the NFκB inhibitor, CAPE, in understanding NMEC infection. StressMarq provides vital research tools to support research into several disciplines. Microglia and astrocytes are also significant sources of C3 in neurodegenerative diseases.
References
- Bacteria reduce flagellin synthesis to evade microglia-astrocyte-driven immunity in the brain, Sun et al., Cell Reports. 2022; 40(1) 111033.

Leave a Reply