Properties
| Storage Buffer | 1X PB pH7.4, 2mM DTT |
| Storage Temperature | -80ºC |
| Shipping Temperature | Dry Ice. Shipping note: Product will be shipped separately from other products purchased in the same order. |
| Purification | Affinity Purified and Size Exclusion |
| Cite This Product | Human Recombinant Tau-441 (2N4R) P301S Mutant Pre-formed Fibrils (CHO-expressed, N-glycosylated) (StressMarq Biosciences Inc., Victoria BC CANADA, Catalog # SPR-516) |
| Certificate of Analysis | Protein certified >95% pure on SDS-PAGE & Nanodrop analysis |
| Other Relevant Information | CHO expression in mammalian cell line may lead to more “human” like phosphorylation/glycosylation patterns. For best results, sonicate immediately prior to use. Refer to the Neurodegenerative Protein Handling Instructions on our website, or the product datasheet for further information. Monomer source is catalog# SPR-515. |
Biological Description
| Alternative Names | MAPT, intracellular neurofibrillary tangles, NFTs, paired helical filaments, PHFs, 2N4R |
| Research Areas | Alzheimer's Disease, Neurodegeneration, Neuroscience, Tangles & Tau |
| Swiss Prot | P10636-8 |
| Scientific Background | Mammalian N-glycosylation is present on CHO-secreted tau 2N4R, which contributes to slower migration on SDS-PAGE than E.coli or Baculovirus/Sf9 expressed tau (1, 2). N-glycosylated tau has been identified in human AD-diseased brains, but not healthy brains, and may precede tau hyperphosphorylation (3, 4). N-glycosylation of Tau has been demonstrated to affect its aggregation propensity (5). The tau P301S mutation is associated with early onset neurodegeneration, and functionally reduces microtubule assembly and stimulates fibril assembly (6, 7). Our CHO-expressed Tau 2N4R P301S will readily form fibrils in the absence of heparin and contains mammalian post-translational modifications that may better mimic tau in human AD-brains. |
| References |
1. Guo et al., 2019. A pathogenic tau fragment compromises microtubules, disrupts insulin signaling and induces the unfolded protein response. Acta Neuropathologica Communications. DOI: 10.1186/s40478-018-0651-9 2. Losev et al., 2020. Differential effects of putative N-glycosylation sites in human Tau on Alzheimer’s disease-related neurodegeneration. Cellular and Molecular Life Sciences. DOI: 10.1007/s00018-020-03643-3 3. Zhang et al., 2020. Integrative glycoproteomics reveals protein N-glycosylation aberrations and glycoproteomic network alterations in Alzheimer’s disease. Sci. Adv. DOI: 10.1126/sciadv.abc5802 4. Liu et al., 2002. Role of glycosylation in hyperphosphorylation of tau in Alzheimer’s disease. FEBS. DOI: 10.1016/S0014-5793(02)02228-7 5. Losev et al., 2019. Novel model of secreted human tau protein reveals the impact of the abnormal N-glycosylation of tau on its aggregation propensity. Sci. Rep. https://doi.org/10.1038/s41598-019-39218-x 6. Bugiani et al., 1999. Frontotemporal Dementia and Corticobasal Degeneration in a Family with a P301S Mutation in Tau. J Neuropathol Exp Neurol. doi: 10.1097/00005072-199906000-00011. 7. Goedert and Crowther, 1999. Effects of frontotemporal dementia FTDP-17 mutations on heparin-induced assembly of tau filaments. FEBS Lett. DOI: 10.1016/s0014-5793(99)00508-7 |
Product Images
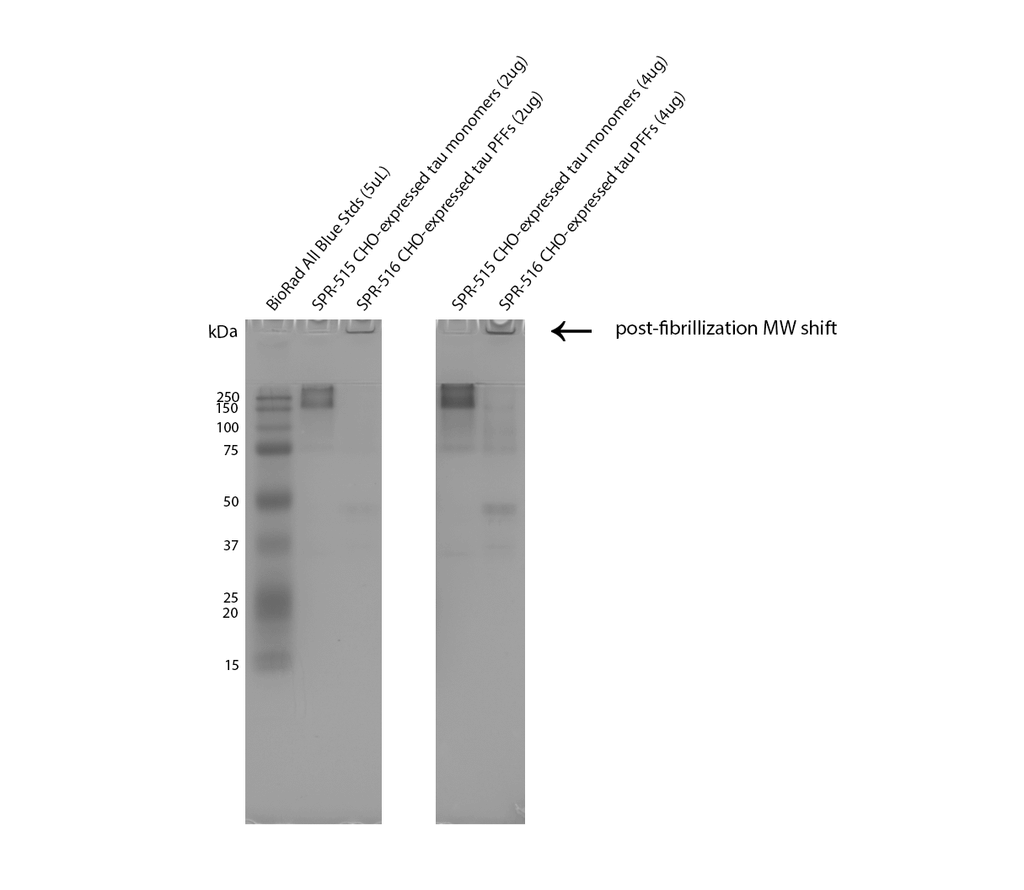
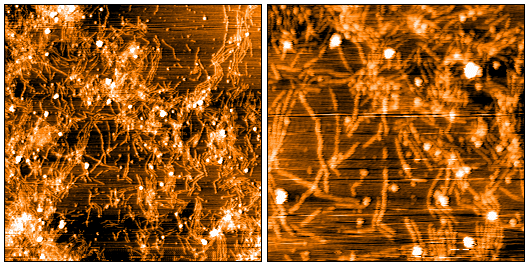
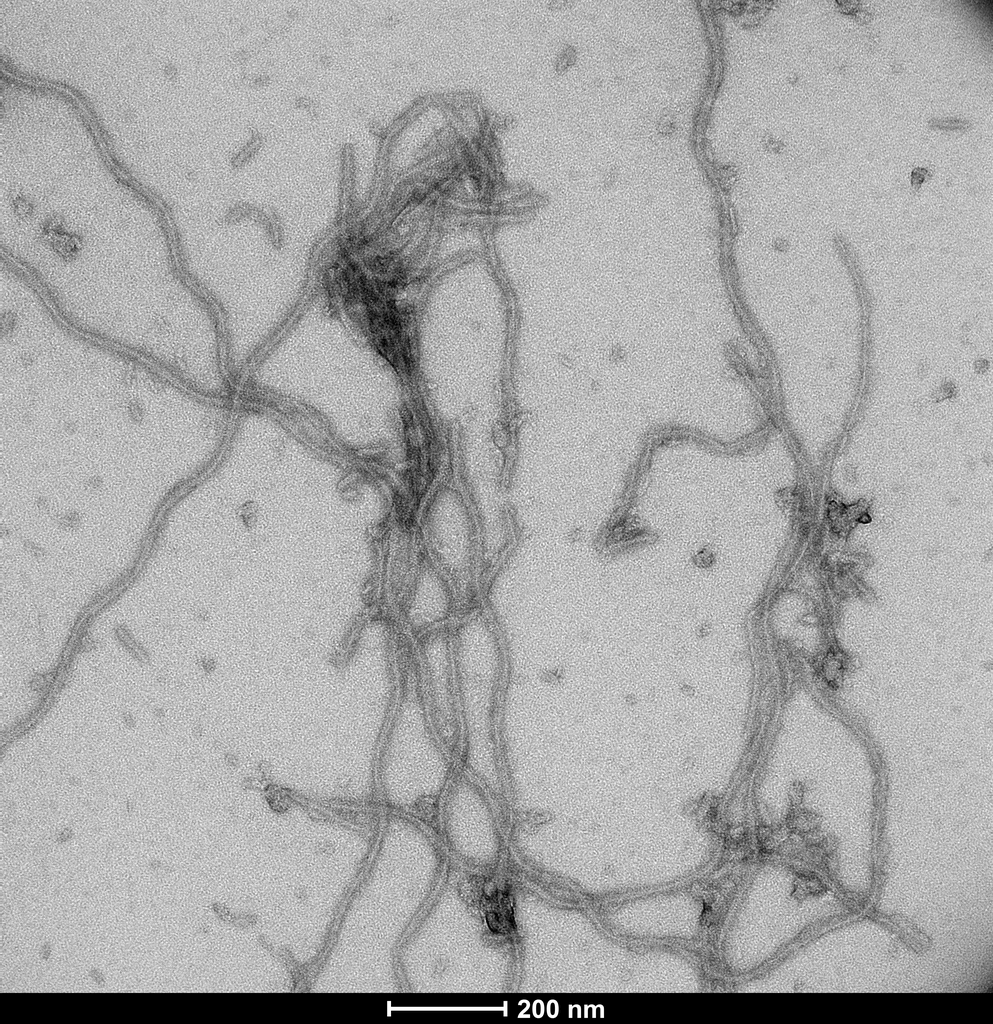
Native-PAGE of CHO-expressed Tau monomers (SPR-515) & fibrils (SPR-516) on a 12% Tris-glycine gel (no SDS). Lane 1: Biorad All Blue Standards (4uL). Lane 2: CHO-expressed hTau 2N4R P301S monomer (2ug). Lane 3: CHO-expressed hTau 2N4R P301S fibril (2ug). Lane 4: n/a. Lane 5: CHO-expressed hTau 2N4R P301S monomer (4ug). Lane 6: CHO-expressed hTau 2N4R P301S fibril (4ug). Note: native proteins typically migrate much slower through a gel compared to reduced, SDS-treated MW standards.
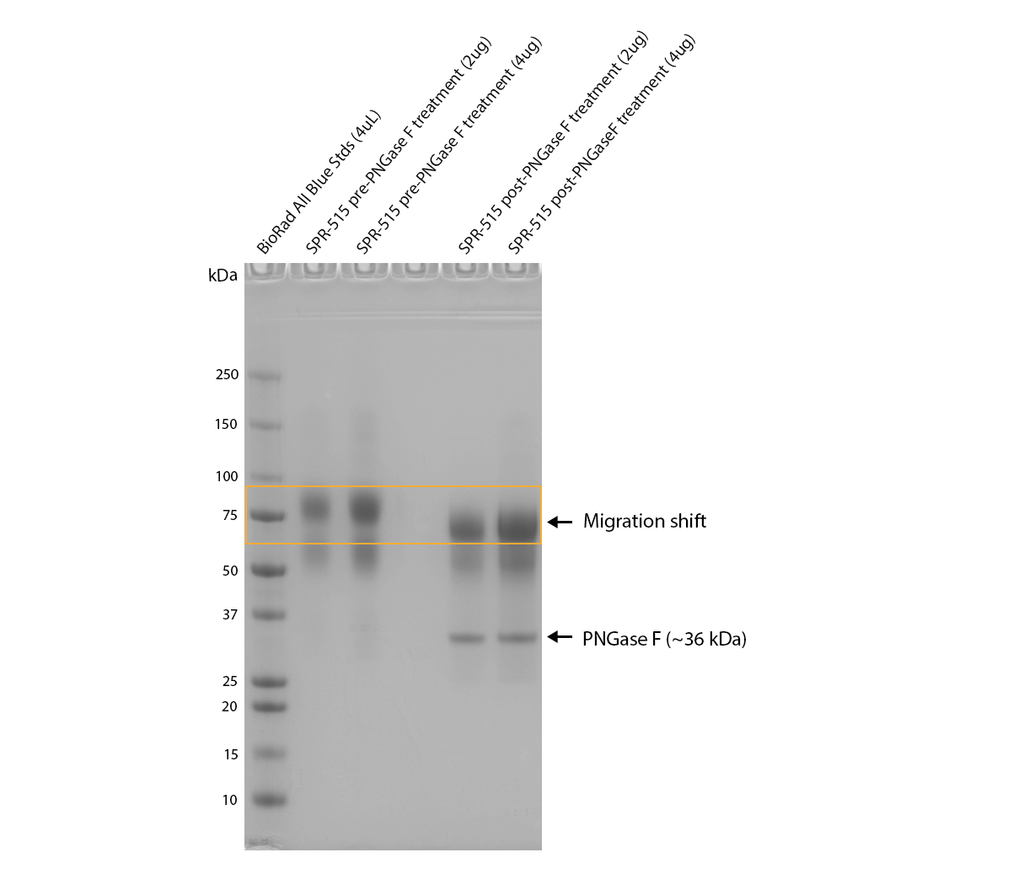
PNGase F treatment of CHO-expressed tau shows an observable shift in apparent MW, indicating the presence of N-glycosylation. Monomers were treated with PNGase F (NEB), a glycosidase which specifically cleaves between the innermost GlcNAc and asparagine residues of N-linked oligosaccharides, and incubated at 37oC for 1 hour and run on a 5-12% gradient Bis-Tris gel.
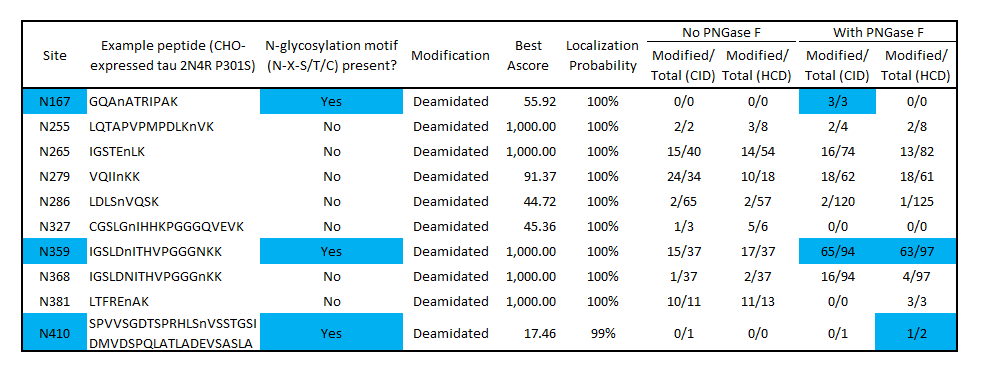
Modified/Total deamidation spectrum counts as determined by mass spectrometry of CHO-expressed tau before and after PNGase F treatment identifies potential N-glycosylation sites at N167, N359 and N410. Blue color indicates deamidation sites that match the N-glycosylation motif (N-X-S/T/C) and have a higher deamidation count after PNGase F treatment. No deamidation was present at N167 or N410 without PNGase F, suggesting these residues are protected from nonspecific deamidation by N-glycosylation. Some deamidation was present at N359 without PNGase F treatment, indicating a population of monomers is not glycosylated at this position. Several non-consensus, non-PNGase F-dependent deamidation sites were present, which may have occurred during production or the mass spectrometry workflow. Both CID and HCD fragmentation methods were used to improve sequence coverage and deamidation detection. Overall protein sequence coverage was 82%, with a localization probability cutoff set at ≥95%.
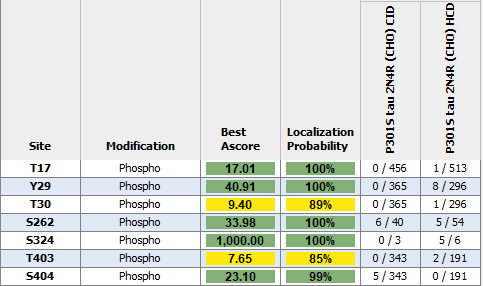
Modified/Total phosphorylation PTM spectrum counts reveal up to 7 phosphorylation sites on human P301S Tau 2N4R monomers expressed using CHO as determined by mass spectrometry. Both CID and HCD fragmentation methods were used to improve sequence coverage and deamidation detection. Protein sequence coverage was 82%. Localization probability cutoff set at ≥80% (yellow) or ≥95% (green). Note: number of phosphorylation sites appear less than Baculovirus/Sf9 expressed tau 2N4R (see StressMarq cat# SPR-471, 472, 496 and 498).
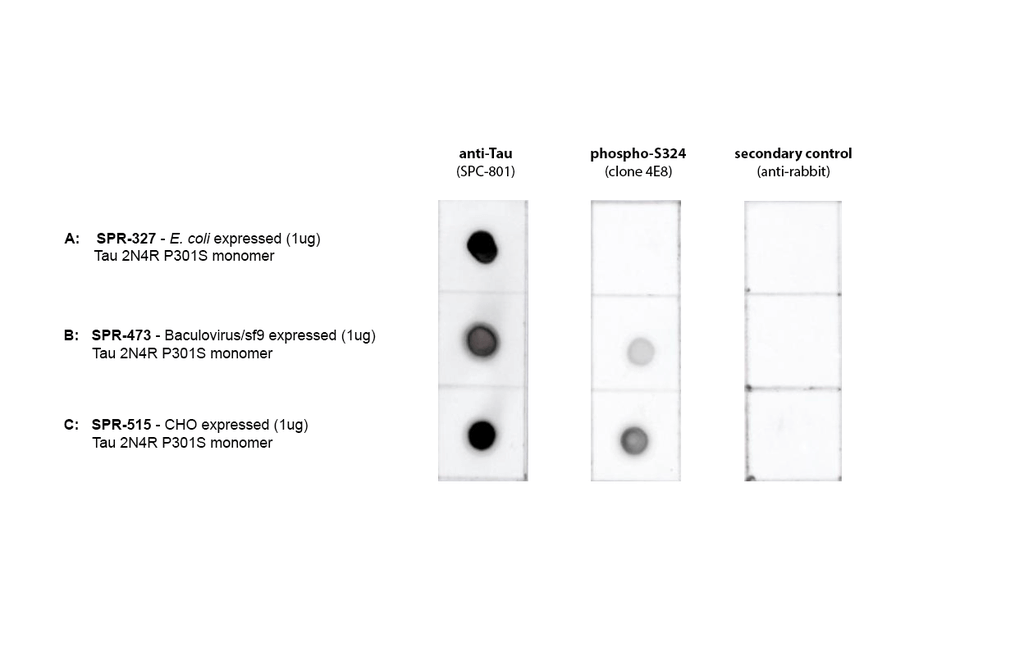
Dot Blot of purified hTau (2N4R) P301S monomers using Stressmarq’s SPC-801 and a phospho-S324 Tau antibody (GeneBio Systems) comparing phosphorylation in E.coli-expressed, baculovirus/sf9-expressed, and CHO-expressed material. Protein was blotted on nitrocellulose, incubated with 1:1000 primary antibodies and/or 1:4000 secondary antibodies. Secondary control is goat-anti rabbit:HRP. Exposed 1 second.

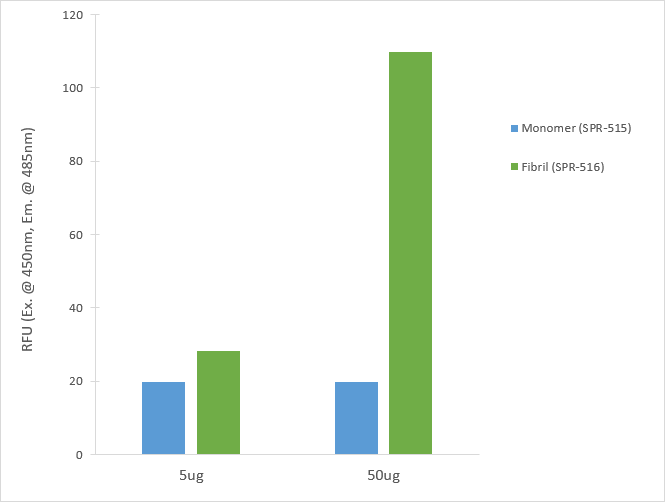
Reviews
There are no reviews yet.